Robust fitting of linear models using Least Absolute Deviations¶
https://en.wikipedia.org/wiki/Least_absolute_deviations
In [18]:
%matplotlib inline
%config IPython.matplotlib.backend = "retina"
import matplotlib.pyplot as plt
from matplotlib import rcParams
rcParams["figure.dpi"] = 300
rcParams["savefig.dpi"] = 300
rcParams['figure.figsize'] = [7, 5]
rcParams["text.usetex"] = True
In [19]:
import tensorflow as tf
import numpy as np
In [20]:
np.random.seed(123)
In [21]:
from lad import lad
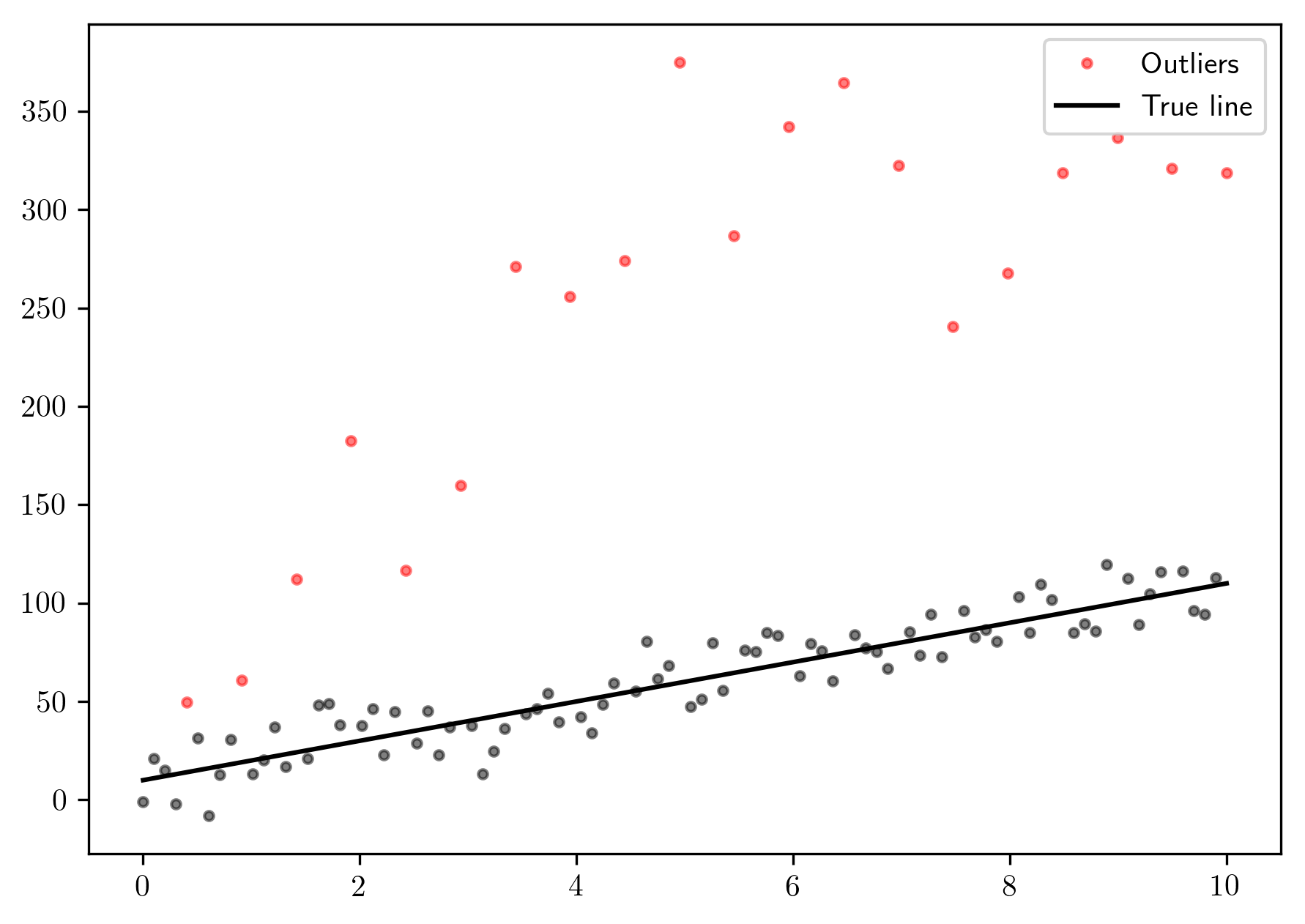
Let’s create some fake data with outliers:
In [22]:
x = np.linspace(0, 10, 100)
noise = 10 * np.random.normal(size=len(x))
y = 10 * x + 10 + noise
In [23]:
mask = np.arange(1, len(x)+1, 1) % 5 == 0
y[mask] = np.linspace(6, 3, len(y[mask])) * y[mask]
In [24]:
plt.plot(x[~mask], y[~mask], 'ok', markersize=3., alpha=.5)
plt.plot(x[mask], y[mask], 'o', markersize=3., color='red', alpha=.5, label='Outliers')
plt.plot(x, 10 * x + 10, 'k', label='True line')
plt.legend()
Out[24]:
<matplotlib.legend.Legend at 0x12c8f2128>
In [25]:
X = np.vstack([x, np.ones(len(x))])
In [26]:
X.shape
Out[26]:
(2, 100)
In [27]:
y.shape
Out[27]:
(100,)
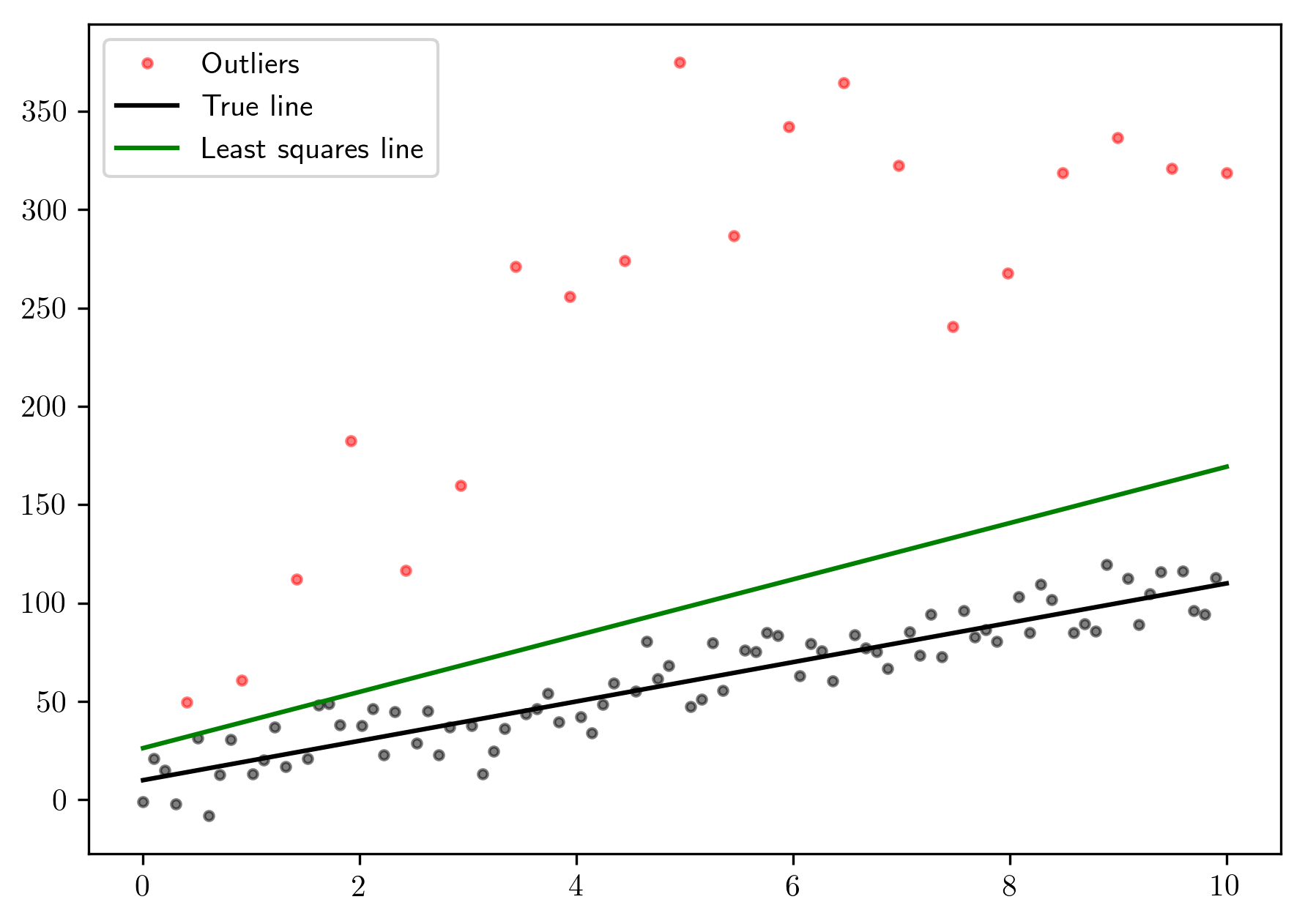
Let’s compute the linear least squares model using
tf.matrix_solve_ls:
In [28]:
with tf.Session() as sess:
X_tensor = tf.convert_to_tensor(X.T, dtype=tf.float64)
y_tensor = tf.reshape(tf.convert_to_tensor(y, dtype=tf.float64), (-1, 1))
coeffs = tf.matrix_solve_ls(X_tensor, y_tensor)
m, b = sess.run(coeffs)
In [29]:
plt.plot(x[~mask], y[~mask], 'ok', markersize=3., alpha=.5)
plt.plot(x[mask], y[mask], 'o', markersize=3., color='red', alpha=.5, label='Outliers')
plt.plot(x, 10 * x + 10, 'k', label="True line")
plt.plot(x, m * x + b, 'g', label="Least squares line")
plt.legend()
Out[29]:
<matplotlib.legend.Legend at 0x12de64e80>
Now let’s use lad to compute the linear model with the least
absolute deviations:
In [30]:
l1coeffs, cov = lad(X.T, y, yerr=np.std(y), cov=True)
In [31]:
sess = tf.Session()
m_l1, b_l1 = sess.run(l1coeffs)
And the estimated values are:
In [32]:
m_l1, b_l1
Out[32]:
(array([ 10.5233137]), array([ 13.6857041]))
The uncertanties on the fitted coefficients are:
In [33]:
unc = np.sqrt(np.diag(sess.run(cov)))
unc
Out[33]:
array([ 0.42935153, 3.5996013 ])
In [34]:
plt.plot(x[~mask], y[~mask], 'ok', markersize=3., alpha=.5)
plt.plot(x[mask], y[mask], 'o', markersize=3., color='red', alpha=.5, label='Outliers')
plt.plot(x, 10 * x + 10, 'k', label="True line")
plt.plot(x, m * x + b, 'g', label="Least squares line")
plt.plot(x, m_l1 * x + b_l1, 'blue', label="Least absolute deviations line")
plt.fill_between(x, (m_l1 - 1.96 * unc[0]) * x + b_l1 - 1.96 * unc[1],
(m_l1 + 1.96 * unc[0]) * x + b_l1 + 1.96 * unc[1], alpha=.5)
plt.legend()
Out[34]:
<matplotlib.legend.Legend at 0x12f80def0>
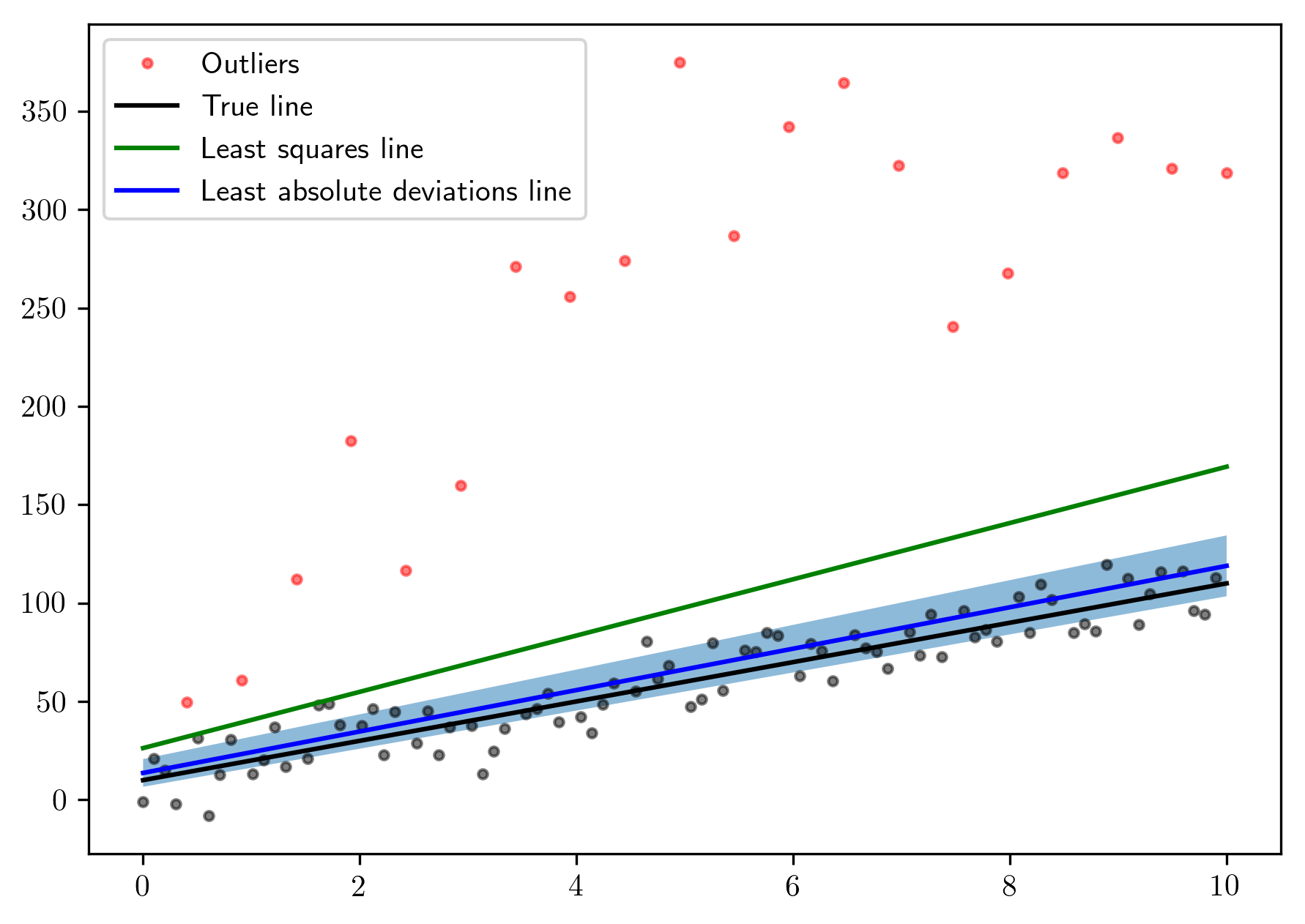
The shaded region is the \(95\%\) confidence interval.